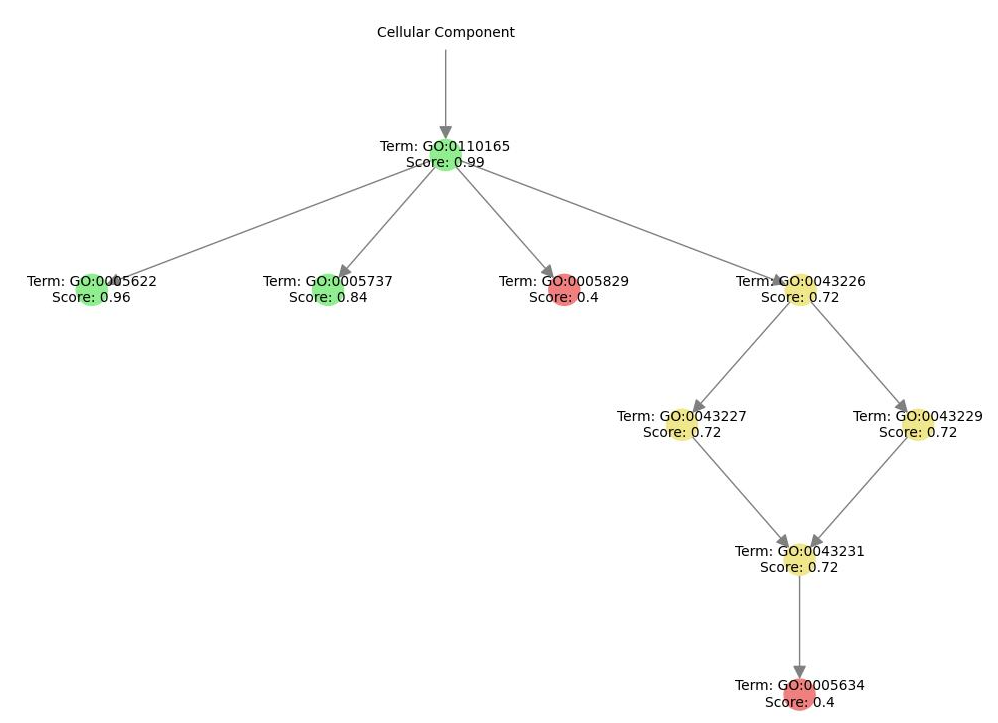
Prediction Page
This page allows users to input their FASTA sequences to predict protein functions.
Input:
Users must enter the following values on the Prediction page:
- Model: This parameter selects the model to be predicted, between SUPERMAGO+Web and SUPERMAGOv2+Web. By default, the selected model is SUPERMAGOv2+Web.
- Minimum Confidence: This parameter specifies the minimum confidence level required for a term to be considered during output generation.
- FASTA Sequences: The sequences to be predicted should be provided in FASTA format. Each query can include up to 50 sequences and is limited to 100,000 characters per task.
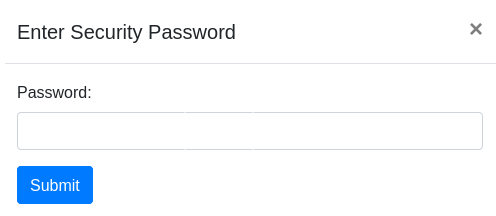
- Add Security Password: Checkbox that determines whether the task will have a password. If not selected, all system users will be able to view the predicted proteins and terms. If the user select the option to add a security password, a unique password for the submitted task will be generated. This password will be required to access the results. You must save the password to access the results.
Errors:
If the input is empty or does not contain sequences in the FASTA format, a notification will appear indicating this error:

Notification for submission with input empty.
If the input has more than 100,000 characters, a notification will appear indicating this error:

Notification for submission with more than 100,000 characters.
Submission:
If no errors are found during the analysis of submission, the task will be added to the queue for execution.
If the user did not select to add a security password, a notification will indicate the task ID:
Notification of task ID if no security password is added.
Otherwise, if the user selected to add a security password, the notification will show the task ID and the password to access the results after processing. You must save the password to access the results:

Notification of task ID and password if a security password is added.
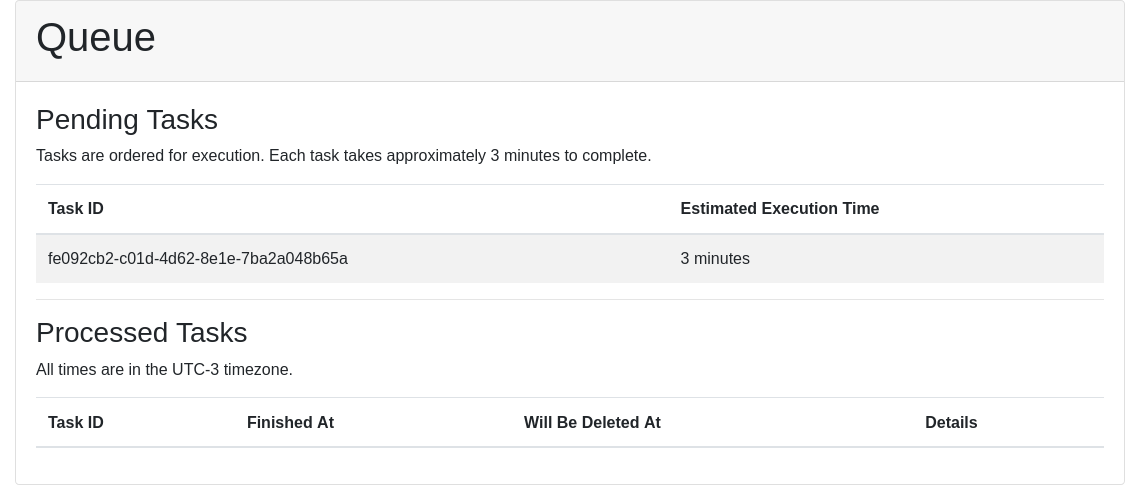
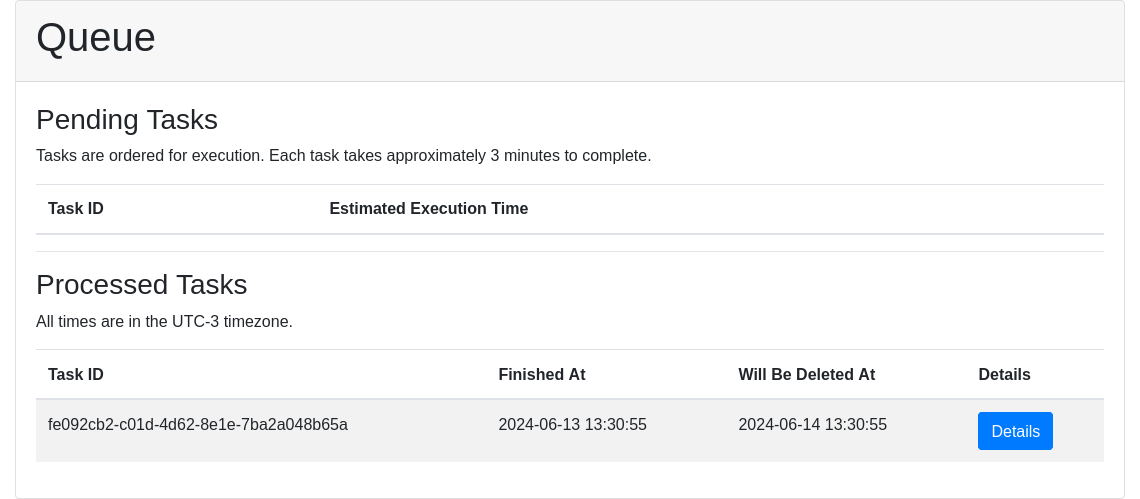
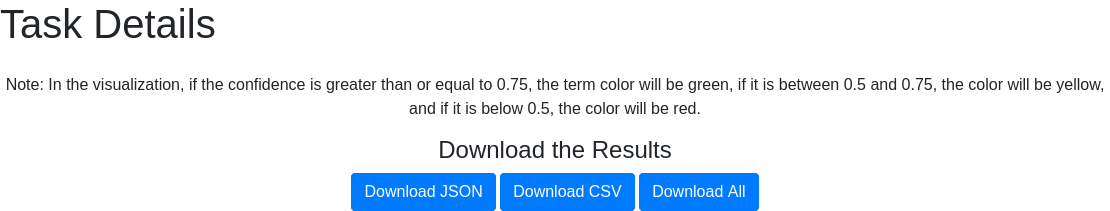
After submission, a window indicates the ID of the submitted task. With this ID, users can track their request on the Queue page.